Leveraging open mobility data from Meta to correct underestimations in disease spread.
A collaboration between Meta (Data for Good) and EV / The Modelers.
Introduction:
The COVID-19 pandemic showcased that the accuracy of epidemic monitoring at a global level can be improved through the inclusion of non-traditional data sources and artificial intelligence. This case study explores how colocation and open mobility data from Meta can ultimately enhance the estimation of the reproduction ratio, which informs public health decision makers about the average number of people who will become infected from contact with an originally infected person. Sorbonne Université, the French National Institute of Health and Medical Research (INSERM) and the Department of Applied Science and Technology (DISAT) at Politecnico di Torino, are leading academic institutions dedicated to advancing scientific research and technological innovation. A team of modelers working across these institutions have been developing data-rich mathematical models to study how infectious diseases spread in human and animal populations. This modeling team reached out to Meta’s Data for Good team to access large-scale mobility data that could improve modeling for COVID-19 and other infectious diseases.
Challenges:
The research team’s work addresses the problem of estimating the reproduction ratio of an infectious disease epidemic, also known as reproductive number, or R. The reproduction ratio is the average number of secondary infections that each case generates, and is a commonly used indicator to monitor and forecast the evolution of an epidemic wave. Accurately estimating R is crucial to inform policies and guide public health interventions. The standard way to estimate the reproduction ratio is to infer it from data from epidemiological surveillance, such as hospitalizations, deaths, testing, or other proxies of infections. However, surveillance data may lead to biased estimates of the reproduction ratio in populations in cities and other geographically distinct communities that are connected through human mobility. Heterogeneities in the contact network shape the way epidemics spread, hiding the true dynamic structure of the epidemic process from population-level surveillance data. Crucially, this means that inferences on surveillance data may either overestimate or underestimate the reproduction ratio over long periods, misinforming authorities and possibly leading to inappropriate responses.
Utilizing Meta Data for Good:
To tackle this challenge, researchers from the Sorbonne University, INSERM and Polytechnic of Turin devised a methodology for inferring the reproduction ratio by combining surveillance data, census data, and colocation and mobility data from Meta’s Data for Good program. The team primarily used Meta colocation maps, which are calculated from mobile phone data and show the proportion of time that residents of different communities within the same country spend in proximity to one another. These datasets provide precise information on the heterogeneous contact patterns that shape the evolution of an infectious epidemic at subnational resolution. The team also used Meta’s open source Movement Range Maps for additional validation and corrections to their modeling. Meta colocation data, together with census data, allowed the team to reconstruct a reproduction operator that reveals the average number of secondary cases each infected individual produces, depending on the region of residence of both the infectious carrier and the newly infected person. After reconstructing the reproduction operator, the research team can evaluate the bias in estimates of the reproduction ratio obtained from population-level surveillance data. Leveraging Meta colocation data to measure close-proximity contact rates within and between communities, the new method can infer the reproduction ratio using only surveillance data with no need to parametrize complex mathematical models describing individual behavior.
Results and Impact:
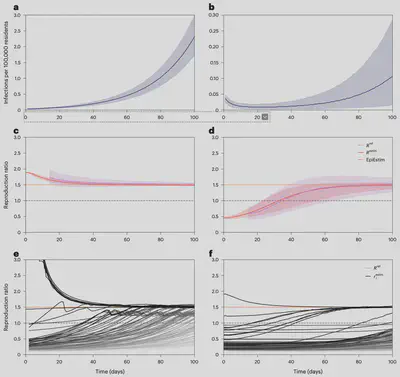
The research team applied their new corrected reproduction ratio retrospectively to the COVID-19 epidemic in France, focusing on the months of January and February 2021. Crucially, this correction suggested that traditional surveillance estimates consistently under-estimated the reproduction ratio during that period, suggesting a subsiding epidemic wave. This estimate was at odds with the actual events, which showcased a growing epidemic wave - ultimately known as the third French wave - and led to the national lockdown enforced on April 3, 2021. The corrected reproduction ratio, on the other hand, would have consistently signaled a growing epidemic wave throughout the first three months of the year. These findings are highly relevant to the debate over the public health response in France at the time. It is conceivable that the underestimation of the third wave’s severity by traditional surveillance could have contributed to delaying the enforcement of stricter movement restrictions, which eventually became inevitable in the spring of 2021.
Future Plans:
The research team’s findings to correct for underestimates in the reproduction ratio could be applied to any epidemic where human mobility is a driver of infections. This includes diseases spread by mosquitoes like dengue that are currently burdening developing countries in the tropics and increasingly present in temperate areas. The team is working with Meta to use their new Activity Space Maps to adapt the methodology to other vector-borne diseases. The researchers from INSERM/Sorbonne U involved in this project also work with the national agency for the modeling of infectious disease outbreaks. This collaboration with the local government will allow them to apply this methodology to emergent and re-emergent epidemic outbreaks, both of COVID-19-like respiratory pathogens and mosquito-borne viruses. To share their work with the wider public health community, the research team has also open sourced their models in an easy-to-use library, built upon existing widely-used frameworks, which is designed to enable correction to existing estimates using human mobility data and surveillance records. These models can be utilized by individuals with basic programming knowledge in Python and do not require field-specific expertise or advanced mathematical skills. The research team plans to develop these tools further to include other data from Meta and to cover additional pathogens for the benefit of disease modelers and public health officials worldwide.
Contact Information:
- INSERM, Sorbonne Université: eugenio.valdano@inserm.fr
- Department of Applied Science and Technology (DISAT) at Politecnico di Torino: michele.refiorentin@polito.it
- EURECAT, Centro Tecnológico de Cataluña: piero.birello@gmail.com
- Meta Data for Good: dataforgood@fb.com
Additional Resources:
- Read the full article in Nature Physics (free-access link: rdcu.be/dFJgn)
- Meta Data for Good